SETH
SNP Extraction Tool for Human Variations
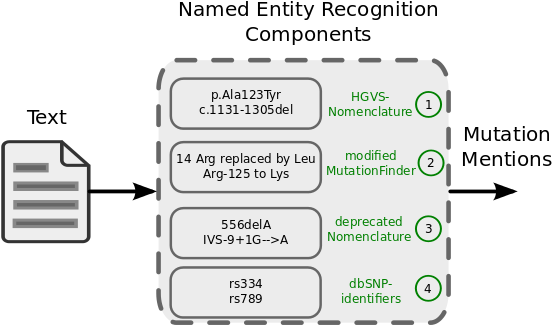
SETH is a software that performs named entity recognition (NER) of genetic variants (with an emphasis on single nucleotide polymorphisms (SNPs) and other short sequence variations) from natural language texts. SETH allows to recognize the following mutation subtypes: substitution, deletion, insertion, duplication, insertion-deletion (insdel), inversion, conversion, translocation, frameshift, short-sequence repeat, and literal dbSNP mention. Recognized mutation mentions can be grounded to the Human Mutation Nomenclature (HGVS) and normalized to dbSNP identifiers or UniProt sequences. For named entity recognition, SETH builds on four individual components:
1.) Mutations following the HGVS nomenclature (den Dunnen and Antonarakis, 2000) are recognized by implementing an Extended Backus–Naur (EBNF) grammar proposed by Laros et al. (2011) using Scala combinators. We modified this grammar to allow the detection of frequently observed deviations from the nomenclature.
2.)To get hold of substitutions not following the nomenclature, SETH integrates MutationFinder (Caporaso et al., 2007). SETH modifies MutationFinder’s original capabilities in order to match a wider scope of substitutions (DNA substitutions, nonsense mutations, and ambiguous mutations) not following the HGVS nomenclature. This is done by modifying the original MutationFinder implementation together with additional and modified regular expressions.
3.) Mutations (substitutions, deletions, insersions, frameshifts, …) not following the HGVS nomenclature, but earlier proposals for a nomenclature, are recognized using a separate set of regular expressions.
4.) Mutations described as literal dbSNP-identifiers are recongized using a regular expression.
Results from the four different components are collected, merged, and represented as the following object MutationMention. The general NER-workflow is also depicted in the following figure.
 .
.
If possible, extracted SNP mentions are linked to dbSNP or UniProt-KB seqeuence. This process is referred to as named entity normalization (NEN). For normalization SETH requires a list of potential entrez gene candidates/identifiers as well as a local dbSNP or UniProt database. Gene names may either come from dedicated gene name recognition and normaluzation tools, such as GNAT. Alternatively, we recommend the use of NCBI’s gene2pubmed database. SETH currently uses these two data-sources but can easily extended with other gene-NER tools.
Cite SETH
SETH detects and normalizes genetic variants in text.
BibTeX
@Article{SETH2016,
Title= {SETH detects and normalizes genetic variants in text.},
Author= {Thomas, Philippe and Rockt{\"{a}}schel, Tim and Hakenberg, J{\"{o}}rg and Lichtblau, Yvonne and Leser, Ulf},
Journal= {Bioinformatics},
Year= {2016},
Month= {Jun},
Doi= {10.1093/bioinformatics/btw234},
Language = {eng},
Medline-pst = {aheadofprint},
Pmid = {27256315},
Url = {http://dx.doi.org/10.1093/bioinformatics/btw234}
}
Text
Thomas, P., Rocktäschel, T., Hakenberg, J., Mayer, L., and Leser, U. (2016). SETH detects and normalizes genetic variants in text. Bioinformatics (2016)
Get SETH
Here we descirbe alternative ways to work with SETH
1.) Download JAR file
Ready-to-use releases available at https://github.com/rockt/SETH/releases/.
2.) Build SETH on your own using Maven:
git clone https://github.com/rockt/SETH.git
cd SETH
mvn clean compile assembly:single
mv ./target/seth-1.4.0-Snapshot-jar-with-dependencies.jar seth.jar
3.) Import in Maven from jitpack
For maven, add a new repository pointing to jitpack.
<repository>
<id>jitpack.io</id>
<url>https://jitpack.io</url>
</repository>
And add the following dependency, and specify the release version
<dependency>
<groupId>com.github.rockt</groupId>
<artifactId>SETH</artifactId>
<version>1.4.0</version>
</dependency>
4.) UIMA wrapper for SETH
In fimda SETH is provided with an UIMA wrapper, as well as integrated web-service provided as docker container. FIMDA is originally developed for the integration of SETH into the openminted platform.
5.) Scalable web-service for SETH
In sia SETH is bundled into a web-service for the Becalm platform.
6.) Docker container for SETH
We also provide Named Entity Recognition facilities using a REST-API and a docker container.
Download docker container from docker-hub
docker pull erechtheus79/seth
docker run -d -p 8080:8080 --network host erechtheus79/seth
Annotate document using GET request
Annotate the string “Two common mutations, c.35delG and L90P accounted for 72.1…”, by sending the string to http://localhost:8080/rest/message/get/.
curl http://localhost:8080/rest/message/get/Two%20common%20mutations,%20c.35delG%20and%20L90P%20accounted%20for%2072.1.
Result:
[
{
"type": "SUBSTITUTION",
"tool": "MUTATIONFINDER",
"location": {
"start": 35,
"stop": 39
},
"text": "L90P",
"ref": null,
"wtResidue": "L",
"mutResidue": "P",
"position": "90",
"nsm": false,
"psm": true,
"ambiguous": false,
"patternId": 25,
"normalized": null,
"transcripts": null,
"start": 35,
"end": 39,
"bestNormalized": null
},
{
"type": "DELETION",
"tool": "SETH",
"location": {
"start": 22,
"stop": 30
},
"text": "c.35delG",
"ref": "c.",
"wtResidue": "G",
"mutResidue": null,
"position": "35",
"nsm": false,
"psm": false,
"ambiguous": false,
"patternId": 0,
"normalized": null,
"transcripts": null,
"start": 22,
"end": 30,
"bestNormalized": null
}
]
7.) Download precomputed PubMed results
Precomputed results are available in GeneView here.
Examples for named entity recognition
Command-line Usage
java -cp seth.jar seth.ner.wrapper.SETHNERAppMut "Causative GJB2 mutations were identified in 31 (15.2%) patients, and two common mutations, c.35delG and L90P (c.269T>C), accounted for 72.1% and 9.8% of GJB2 disease alleles."
MutationMention [span=91-99, location=35, wtResidue=G, text=c.35delG, type=DELETION, tool=SETH]
MutationMention [span=104-108, mutResidue=P, location=90, wtResidue=L, text=L90P, type=SUBSTITUTION, tool=MUTATIONFINDER]
MutationMention [span=110-118, mutResidue=C, location=269, wtResidue=T, text=c.269T>C, type=SUBSTITUTION, tool=SETH]
java -cp seth.jar seth.ner.wrapper.SETHNERAppMut "G-banding and spectral karyotyping showed 46,XX,t(9;11)(p22;p15)."
MutationMention [span=42-64, text=46,XX,t(9;11)(p22;p15), type=COPY_NUMBER_VARIATION, tool=SETH]
Examples for named entity normalization
Given mutation mentions and a list of potential genes (i.e. Entrez gene identifiers), SETH normalizes SNPs to dbSNP identifiers. To extract gene mentions, we use the output of the tool GNAT (Hakenberg et al., 2011) together with the gene2pubmed information from NCBI. Parts of the dbSNP database have to be locally installed for speeding up the normalization process. For user convenience, we provide a dbSNP137 as embedded Derby database (~2GB). We also provide a derby database for a more recent human dbSNP dump dbSNP147. Please be warned that the download is 7.5GB compressed and requires 51 GB uncompressed space. Runtime requirements for normalization also substantially increases with this version of dbSNP in comparison to the smaller dump. For example, normalization of the 296 documents from Thomas et al. (2011) increases from approximately 30 seconds to 140 seconds on a commodity laptop. We highly encourage the use of an dedicated database, such as MySQL or PostgreSQL to increase runtime. Please note, that this derby database dump contains only human data (UniProt, dbSNP, GNAT, gene2Pubmed). Otherwise, the resulting derby database becomes too large for distribution. At the end of this readme we describe the process to generate the derby database. Feel free to contact us if you observe any problems, or if you would like to host database dumps for species other than human.
Command-line Usage
To use SETH’s NEN component from the command line, you need to provide a XML property file that handles the connection to the Derby database. Subsequently, you can provide a tab-seperated file (with PubMed ID, mutation mention, start- and end-position) SETH should normalize to dbSNP (i.e. rs numbers).
java -cp seth.jar de.hu.berlin.wbi.process.Normalize resources/property.xml resources/snpExample.txt
Normalising mutations from 'resources/snpExample.txt' and properties from 'resources/property.xml'
16 mutations for normalisation loaded
PMID Mutation Start End dbSNP
15345705 G1651A 419 425
15290009 V158M 149 158 rs4680
15645182 A72S 15 23 rs6267
14973783 V103I 208 213 rs2229616
15457404 V158M 937 946 rs4680
15098000 V158M 349 358 rs4680
12600718 R219K 599 604 rs2230806
12600718 R1587K 945 951 rs2230808
15245581 T321C 1356 1361 rs3747174
15464268 S12T 621 625 rs1937
11933203 C707T 685 691
14984467 C17948T 577 585
15268889 G2014A 1316 1322
15670788 C2757G 726 733
15670788 C5748T 755 762
15564288 R72P 1204 1212 rs1042522
15564288 K751Q 747 756 rs13181
15564288 D312N 722 731 rs1799793
14755442 E2578G 699 709 rs1009382
15615772 P141L 1040 1045 rs2227564
Normalization possible for 14/20 mentions
Code Example
SETH allows simple integration into your Java-Projects. A complete pipeline performing all steps (NER+NEN) can be found here: Java-Code
Reproducing our results for NER and NEN
Evaluate NER
SETH corpus (630 abstracts)
java -cp seth.jar seth.seth.eval.ApplyNER resources/SETH-corpus/corpus.txt resources/mutations.txt false resources/SETH-corpus.seth
java -cp seth.jar seth.seth.eval.EvaluateNER resources/SETH-corpus.seth resources/SETH-corpus/yearMapping.txt resources/SETH-corpus/annotations/
Precision 0.98 Recall 0.86 F₁ 0.91
MutationFinder-development corpus using original MutationFinder evaluation scripts (Caporaso et al., 2007)
java -cp seth.jar seth.seth.eval.ApplyNER resources/mutationfinder/corpus/devo_text.txt resources/mutations.txt true resources/devo_text.seth
python resources/mutationfinder/origDist/performance.py resources/devo_text.seth resources/mutationfinder/corpus/devo_gold_std.txt
Precision 0.98 Recall 0.83 F₁ 0.90
MutationFinder-test corpus using original MutationFinder evaluation scripts (Caporaso et al., 2007)
java -cp seth.jar seth.seth.eval.ApplyNER resources/mutationfinder/corpus/test_text.txt resources/mutations.txt true resources/test_text.seth
python resources/mutationfinder/origDist/performance.py resources/test_text.seth resources/mutationfinder/corpus/test_gold_std.txt
Precision 0.98 Recall 0.82 F₁ 0.89
Corpus of Wei et al. (2013); train
java -cp seth.jar seth.seth.eval.ApplyNERToWei resources/Wei2013/train.txt resources/mutations.txt resources/Wei2013.seth
java -cp seth.jar seth.seth.eval.EvaluateWei resources/Wei2013/train.txt resources/Wei2013.seth
Precision 0.93 Recall 0.80 F₁ 0.86
Corpus of Wei et al. (2013); test
java -cp seth.jar seth.seth.eval.ApplyNERToWei resources/Wei2013/test.txt resources/mutations.txt resources/Wei2013.seth
java -cp seth.jar seth.seth.eval.EvaluateWei resources/Wei2013/test.txt resources/Wei2013.seth
Precision 0.95 Recall 0.77 F₁ 0.85
Corpus of Verspoor et al. (2013)
java -cp seth.jar seth.seth.eval.ApplyNerToVerspoor resources/Verspoor2013/corpus/ resources/mutations.txt resources/Verspoor2013.seth
java -cp seth.jar seth.seth.eval.EvaluateVerspoor resources/Verspoor2013/annotations/ resources/Verspoor2013.seth
Precision 0.87 Recall 0.14 F₁ 0.24
Evaluate NEN
Corpus of Thomas et al. (2011)
java -cp seth.jar de.hu.berlin.wbi.process.Evaluate myProperty.xml resources/thomas2011/corpus.txt
Precision 0.96 Recall 0.57 F₁ 0.72 Details: TP 303; FP 14; FN 224
Corpus of OSIRIS (Furlong et al., 2008)
java -cp seth.jar de.hu.berlin.wbi.process.osiris.EvaluateOsiris myProperty.xml resources/OSIRIS/corpus.xml
Precision 0.94 Recall 0.69 F₁ 0.79 Details: TP 179; FP 11; FN 79
References
- Caporaso, J. G. et al. (2007). MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics, 23(14), 1862–1865.
- den Dunnen, J. T. and Antonarakis, S. E. (2000). Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Human Mutat, 15(1), 7–12.
- Furlong, L. I. et al. (2008). Osirisv1.2: a named entity recognition system for sequence variants of genes in biomedical literature. BMC Bioinformatics, 9, 84.
- Hakenberg, J. et al. (2011). The GNAT library for local and remote gene mention normalization. Bioinformatics 27(19):2769-71
- Laros, J. F. J. et al. (2011). A formalized description of the standard human variant nomenclature in Extended Backus-Naur Form. BMC bioinformatics, 12 Suppl 4(Suppl 4), S5.
- Thomas, P. E. et al. (2011). Challenges in the association of human single nucleotide polymorphism mentions with unique database identifiers. BMC Bioinformatics, 12 Suppl 4, S4.
- Verspoor, K. et al. (2013). Annotating the biomedical literature for the human variome. Database (Oxford).
- Wei, C.-H. et al. (2013). tmvar: a text mining approach for extracting sequence variants in biomedical literature. Bioinformatics, 29(11), 1433–1439.
- Yepes, A. J. and Verspoor, K. Mutation extraction tools can be combined for robust recognition of genetic variants in the literature F1000Research 2014, 3:18
Rebuilding the database for SNP normalization
This database is only required for normalization to either dbSNP or UniProt. The following steps are needed if you want to build the database from scratch. We provide an (old/ancient/prehistoric) stand-alone (embedded) Derby database. The import script is tailored towards a PostgreSQL-database, but theoretically any other database can be used. We would be happy to get feedback about using SETH with other databases.
Download the necessary files
Please be aware that downloading the necessary files takes substantial amounts of time and space. The script was last tested with build 153 on 3rd of June 2021 and total download size was approx 200 GB.
1.) Download XML or JSON dump from dbSNP
First, we download the relevant files from dbSNP (either XML or JSON). We can either choose XML or JSON as input format. XML is much smaller in download-size, faster to process, produces a smaller database, but has inferior normalization performance. In general, precision is worse in XML and recall is comparable.
mkdir XML
wget --continue --directory-prefix=XML/ ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/XML/ds\*.gz
mkdir JSON
wget --continue --directory-prefix=JSON/ ftp://ftp.ncbi.nlm.nih.gov/snp/redesign/latest_release/JSON/refsnp-chr\*.bz2
2.1) Download gene2pubmed links from NCBI-Entrez gene
Creates a directory entrezGene and stores data into this folder.
mkdir entrezGene
wget --directory-prefix=entrezGene/ ftp://ftp.ncbi.nih.gov/gene/DATA/gene2pubmed.gz
gunzip entrezGene/gene2pubmed.gz
2.2) Download gene-NER results from the PubTatorCentral repository
Creates a directory pubtator and stores data into this folder.
mkdir pubtator
wget --directory-prefix=pubtator/ https://ftp.ncbi.nlm.nih.gov/pub/lu/PubTatorCentral/gene2pubtatorcentral.gz
gunzip pubtator/gene2pubtatorcentral.gz
3.) Download UniProt-KB and Id-Mapping
Creates a directory uniProt and stores data into this folder.
mkdir uniProt
wget --directory-prefix=uniProt/ ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/complete/uniprot_sprot.xml.gz
wget --directory-prefix=uniProt/ ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/idmapping.dat.gz
4.) Check compression
We identify corrupted files by testing the compression.
Please delete and re-initiate download for all corrupted files.
Corrupt files can be identified by an error similar to:
gzip: ./dbSNP/ds_ch18.xml.gz: invalid compressed data--format violated
find XML/ -name *.gz -exec gunzip --test {} \;
find uniProt/ -name *.gz -exec gunzip --test {} \;
find JSON/ -name *.bz2 -exec bunzip2 --test {} \;
Convert dbSNP-, UniProt-, Entrez-dumps into TSV-files
1.) Parse dbSNP dump (either XML or JSON-files)
Requires as input the file paths with all dbSNP XML (or JSON) files. Parsing took 255 minutes for XML and 3,821 minutes for JSON. The result is ~5GB for XML and 62GB for JSON.
mkdir Out_XML/
time java -cp seth.jar -Djdk.xml.totalEntitySizeLimit=0 -DentityExpansionLimit=0 de.hu.berlin.wbi.stuff.dbSNPParser.xml.ParseXMLToFile XML/ Out_XML/
mkdir Out_JSON/
time java -cp seth.jar de.hu.berlin.wbi.stuff.dbSNPParser.json.ParseJSONToFile JSON/ Out_JSON/ --noRefSeq
Parse UniProt-XML for protein-sequence mutations (PSM) and post-translational modifications (e.g. signaling peptides) (16m processing time)
Requires as input the UniProt-KB dump (uniprot_sprot.xml.gz) and the mapping from Entrez to UniProt (idmapping.dat.gz). Produces uniprot.dat and PSM.dat files at specified location.
java -cp seth.jar seth.Uniprot2Tab uniProt/uniprot_sprot.xml.gz uniProt/idmapping.dat.gz uniProt/uniprot.dat uniProt/PSM.dat
Some postprocessing steps, to ensure that the import data is unique
time sort -u uniProt/uniprot.dat -o uniProt/uniprot.dat #Make uniprot-data unique
time sort -u Out_XML/hgvs.tsv -o Out_XML/hgvs.tsv
time sort Out_XML/PSM.tsv uniProt/PSM.dat -u -o Out_XML/PSM.tsv
time grep -v "-" Out_XML/PSM.tsv > foo.bar && mv foo.bar Out_XML/PSM.tsv
time sort Out_JSON/PSM.tsv uniProt/PSM.dat -u -o Out_JSON/PSM.tsv
time grep -v "-" Out_JSON/PSM.tsv > foo.bar && mv foo.bar Out_JSON/PSM.tsv
Set up PostgreSQL (Docker container)
Please set variables $DatabasePassword and $importData accordingly.
DatabasePassword=yourPwd
importData=yourPath
docker pull postgres
--mount type=volume,source=$HOME/docker/volumes/postgres,target=/var/lib/postgresql/data
docker run --name pg-docker -e POSTGRES_PASSWORD=${DatabasePassword} -d -p 5432:5432 --mount type=bind,source=${importData},target=/var/lib/importData,readonly postgres
Create the database with all necessary tables
createdb -h localhost -U postgres dbsnp-xml
psql -h localhost -U postgres -d dbsnp-xml < SETHDirectory/resources/table.sql
createdb -h localhost -U postgres dbsnp-json
psql -h localhost -U postgres -d dbsnp-json < SETHDirectory/resources/table.sql
Import gene2pubmed, UniProt, PSM, mergeItems, and gene2pubmed into the Database
psql -h localhost -U postgres -d dbsnp-xml
COPY psm FROM '/var/lib/importData/Out_XML/PSM.tsv' DELIMITER E'\t';
COPY uniprot FROM '/var/lib/importData/uniProt/uniprot.dat' DELIMITER E'\t';
COPY hgvs FROM '/var/lib/importData/Out_XML/hgvs.tsv' DELIMITER E'\t';
COPY mergeItems FROM '/var/lib/importData/Out_XML/mergeItems.tsv' DELIMITER E'\t';
COPY gene2pubmed FROM PROGRAM 'tail -n +2 /var/lib/importData/entrezGene/gene2pubmed' DELIMITER E'\t';
DELETE FROM gene2pubmed where taxid != 9606;
psql -h localhost -U postgres -d dbsnp-json
COPY psm FROM '/var/lib/importData/Out_JSON/PSM.tsv' DELIMITER E'\t';
COPY uniprot FROM '/var/lib/importData/uniProt/uniprot.dat' DELIMITER E'\t';
COPY hgvs FROM '/var/lib/importData/Out_JSON/hgvs.tsv' DELIMITER E'\t';
COPY gene2pubmed FROM PROGRAM 'tail -n +2 /var/lib/importData/entrezGene/gene2pubmed' DELIMITER E'\t';
DELETE FROM gene2pubmed where taxid != 9606;
-- COPY mergeItems FROM '/var/lib/importData/Out_JSON/mergeItems.tsv' DELIMITER E'\t';
Import gene2pubmed, UniProt, PSM, mergeItems, and gene2pubmed into the Database
COPY gene2pubtatorcentral FROM '/var/lib/importData/pubtator/gene2pubtatorcentral' DELIMITER E'\t';
-- Remove all entries, where the gene-ID is unknown
DELETE FROM gene2pubtatorcentral WHERE geneIDText = 'None';
-- Add an array covering all entrez-gene ID's
ALTER TABLE gene2pubtatorcentral ADD COLUMN geneID BIGINT[];
UPDATE gene2pubtatorcentral SET geneID = string_to_array(geneIDText, ';'):: bigint[];
CREATE INDEX id_gene2pubtatorcentral ON gene2pubtatorcentral USING GIN (geneId) ;
ALTER TABLE gene2pubtatorcentral DROP geneIDText;
-- Add an array covering all tools
CREATE TYPE datasources AS ENUM ('BioGRID', 'CRAFT', 'CTD', 'gene2pubmed', 'gene_interactions', 'generifs_basic', 'GNormPlus', 'GWAS', 'HPRD', 'MESH', 'MGI', 'RGD', 'TAIR', 'ZFIN');
ALTER TABLE gene2pubtatorcentral ADD COLUMN tools datasources[];
UPDATE gene2pubtatorcentral SET tools = string_to_array(toolText, '|')::datasources[];
CREATE INDEX tools_gene2pubtatorcentral ON gene2pubtatorcentral (tools) ;
ALTER TABLE gene2pubtatorcentral DROP toolText;
VACUUM FULL;
Backup database and restore
pg_dump -h localhost -U postgres dbsnp-xml > dbsnp-xml.sql
lzma --best dbsnp-xml.sql
createdb -h localhost -U postgres dbsnp-xml
lzcat dbsnp-xml.sql.lzma | psql -h localhost -U postgres dbsnp-xml
Derby database from 18th May 2016
We now also provide a derby database for the human dbSNP dump dbSNP147. We highly encourage the use of a dedicated database, such as PostgreSQL to increase runtime. You can build your own database by following the steps explained above (Rebuilding the database for SNP normalization). Please be warned that the derby database is 7.5GB compressed and requires 51 GB uncompressed space. Runtime requirements for normalization also substantially increases with this version of dbSNP in comparison to previous versions. For example, normalization of the 296 documents from Thomas et al. (2011) increases from approximately 30 seconds to 140 seconds on a commodity laptop. Performance of this model on the previously introduced normalization corpora Osiris and Thomas et al. (2011):
| Corpus | Precision | Recall | F₁ |
|---|---|---|---|
| Thomas et al. (2011) | 0.89 | 0.59 | 0.71 |
| Osiris | 0.94 | 0.69 | 0.79 |
Bug reports
Issues and feature requests can be filed online
Contact
For questions and remarks please contact Philippe Thomas. For more details https://www.dfki.de/en/web/about-us/employee/person/phth01/.
